Clinical evidence that drives the development of our innovative AI technology

Objective
To evaluate the performance of a deep learning (DL) system for automated calliper placement to obtain 6 key sonographic measurements of the fetal brain (transventricular [TV] and transcerebellar [TC] planes).
Methods
From 3 centres (2 tertiary referral centres, 1 routine imaging centre), 1497 (583 pregnancies) TV, and 596 (187 pregnancies) TC plane images were obtained retrospectively using 3 commercial ultrasound devices (GE Voluson E8, S10, P8). The calliper positions (X and Y coordinates) for 6 measurements (TV plane: biparietal diameter [BPD], occipitofrontal diameter [OFD], atrial width [AW]; TC plane: transcerebellar diameter [TCD], cisterna magna size [CMS], nuchal fold thickness [NFT]) provided by fetal medicine specialists (FMS) were used as the gold standard. For each measurement, we trained (1200 images/measurement) a DL system (high-resolution network [HR-Net]) to automatically predict the calliper positions (2 per measurement) using the gold standard dataset, and measurements were computed as the Euclidean distance between them. We assessed the performance (calliper position, measurement) of the DL system (vs. 2 FMS) on an independent (unseen) test set of 145 images (145 pregnancies) by computing the mean Euclidean error (DL system vs. 2 FMS) and the absolute agreement (intraclass correlation coefficients [ICC]; two-way random-effects, average rater) for each measurement.
Results
For all 6 measurements, the Euclidean errors (means) were always less than 2.11±0.98mm, and the DL system was in a good (NFT, CMS; ICC > 0.80) to excellent (BPD, OFD, TCD, AW; ICC > 0.90) agreement with 2 FMS.
Conclusion
The successful clinical translation of the proposed DL system is of high value for training novice users and in low-resource settings that lack well-trained specialists for obtaining reliable fetal structural measurements.

The dotted red lines in each graph represent the 95% Confidence Interval (CI)


Aim
To develop and validate an artificial intelligence system (AIS) to automatically obtain 9 key fetal brain measurements.
Methods
A total of 2435 2D- ultrasound images of transventricular (TV) and transcerebellar (TC) planes were retrospectively obtained from 582 subjects (targeted mid trimester assessment; 3 centres) using 3 ultrasound devices (GE Voluson E8/P8/S10) to train and test a (dataset split = 80:20) a custom AI model (U - Net) to segment 10 fetal brain structures. On an independent test set (144 images; 1 per subject), using the segmentation masks, 9 measurements (biparietal diameter [BPD], occipitofrontal diameter [OFD], cephalic index [CI], head circumference [HC], atrial width [AW], cavum septum pellucidum [CSP] ratio, transcerebellar diameter [TCD], cisterna magna size [CMS], Nuchal Fold Thickness [NFT]) were computed and Benchmarked (intraclass correlation coefficients [ICC], mean error) against the manual measurements of 4 fetal medicine specialists [FMS].
Results
The AIS offered a good segmentation performance (mean Dice coefficient: 0.83). When compared to the 4 FMS, the automated measurements were in excellent (BPD: 0.99, OFD:0.95, HC: 0.98),good (CI: 0.72,TCD: 0.89), and moderate agreements (CSP ratio: 0.51, AW:0.57, CMS: 0.65, NFT: 0.68). The mean intra-rater differences for each FMS were comparable to the absolute error between the AIS and FMS panel.
Conclusions
The proposed AI system can assist novice users in delivering standardized quality prenatal examinations in high volume settings.

Background
The ‘lemon sign' refers to the inward scalloping of the frontal bones in a fetal skull and has a strong clinical association with multiple anomalies such as open neural tube defects, encephalocele, etc. The automated detection of lemon sign from ultrasonography (USG) scans can assist novice sonographers and clinicians in low-resource settings in providing timely and informed referrals to tertiary/specialist centers for further examinations. In this study, we design and validate a fully automated artificial intelligence (AI) system to detect lemon sign from 2D USG images of the fetal brain.
Methods
A total of 5791 USG images (normal/lemon sign cranium:4710/1081) of the transventricular (TV) and transcerebellar (TC) planes were retrospectively obtained from 1192 pregnancies (lemon sign: 44 pregnancies) through targeted mid-trimester USG examination at 2 tertiary referral centers using 3 commercially available USG devices (General Electric [GE] Healthcare; GE Voluson E8/P8/S10). We developed two AI networks to (1) identify the fetal cranium and obtain segmentation masks; (2) classify the segmentation masks as a lemon sign or normal. A U-Net based cranium segmentation network was trained and tested on 2400 and 719 images respectively. 'Enriched cranium segmentation masks’ (segmentation masks multiplied with latent space feature maps) were extracted for the remaining 2672 USG images using the trained segmentation network. A classifier network was trained and tested (equal number of lemon sign and normal cases) on the 800 and 1872 enriched cranium segmentation masks, respectively. The Dice coefficient was used to evaluate the performance of the cranium segmentation network (scale = 0: no-overlap; 1.0: complete overlap; comparison against manual segmentations). The sensitivity, specificity, and area under the receiver operating characteristics curve (AUC) were used for evaluating the performance of the classifier network. We also used GradCam maps to qualitatively analyze the important regions focused by the classifier to detect a lemon sign cranium and offer clinically interpretability of the AI network.
Results
The segmentation network achieved a Dice coefficient of 0.82. The normal/lemon sign classifier offered a sensitivity, specificity, and AUC of 0.88, 0.99, and 0.99. Qualitative analysis of the GradCam maps confirmed that the classifier network focused on the inward scalloping of the frontal bones in most cases.
Conclusion
The proposed AI system offers clinically interpretability and good performance in the fully automated detection of lemon sign fetal craniums. Its clinical translation to low-resource/remote settings can help sonographers provide timely referrals to specialists for detailed evaluation and management.
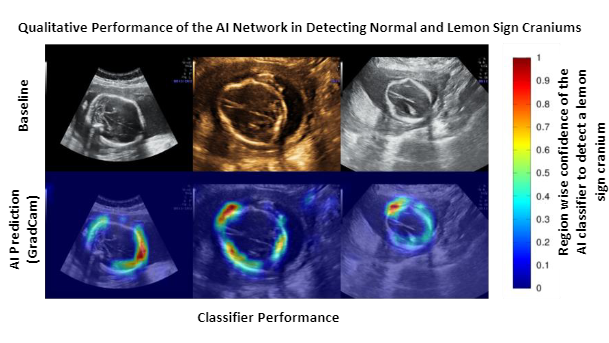
The second row represents the GradCam maps of the classifier for the baseline images.
The color scale on the right, indicates the region-specific confidence given by the classifier
(Blue = low confidence; Red = highest confidence) to detect lemon sign craniums.

Objective
To validate the clinical and workflow benefits of Origin Health Examination Assistant (OHEA), an artificial intelligence (AI) based system capable of semi-automated caliper placement and measurement of multiple fetal brain structures.
Methods
Origin Health Examination Assistant (OHEA) is an AI based assistive software system consisting of multiple AI algorithms (one per measurement) for the semi-automated (adjustable by the user) caliper placement to compute 11 key measurements from fetal brain mid-trimester (18-24 weeks) examinations. All AI algorithms were trained and clinically validated on an expert annotated dataset of 1,650 (1,150 patients) and 300 (280 patients) 2D ultrasound images of the fetal brain axial (transventricular and transcerebellar) and sagittal (mid-sagittal) views obtained from a single tertiary fetal care centre.
The OHEA’s caliper placement (definitions based on Fetal Medicine Foundation guidelines) across fetal brain axial and sagittal views include the biparietal diameter (BPD), occipitofrontal diameter (OFD), atrial width of the lateral ventricle (AW), nuchal fold thickness (NFT), cisterna magna size (CMS), transcerebellar diameter (TCD), corpus callosum length (CC-L), corpus callosum body with (CC-B), vermis anteroposterior diameter (V-AP), and vermis superioinferior (V-SI) diameter. To assess the clinical and workflow benefits of OHEA, we retrospectively obtained an external test set of 358 examinations (1 per patient) from a single tertiary fetal care centre between July 2019 and February 2022. We benchmarked the performance against a reader panel of 7 clinicians (6 OBGYNs and 1 Radiologist) trained in fetal medicine.
A set of 110 images (110 patients; 93 axial and 17 sagittal images) consisting of normal (96 cases) and abnormal cases (14 cases; 1 enlarged cisterna magna, 3 increased nuchal fold thickness, 6 choroid plexus cyst, 1 agenesis of the corpus callosum, 3 partial agenesis of the corpus callosum, and 1 vermian hypoplasia) that were deemed optimal by 2 senior fetal medicine trained clinicians were used for the study. In the first phase of the study, to establish a baseline, all readers manually placed the caliper points and measured as in current clinical practice for all the 11 measurements on all the 110 images. In the second phase of the study, the entire process was repeated with the OHEA placing the caliper points and computing the measurements, while the readers were given the option to adjust if required.
We obtained the intraclass correlation coefficient (ICC; two-way random, mean of k raters, and absolute agreement) and assessed the agreement in measurements between the reader panel and OHEA. We also assessed the inter-rater variability among the reader panel (absolute error rates in measurements), time taken (caliper-positioning and adjustment), and the number of keystrokes for both phases of the study (with and without the use of OHEA).
Results
On average, we observed that the ICC between the panel and OHEA for all the measurements were 0.963 indicating excellent agreement. Specifically, the ICC for individual measurements were 0.993 (BPD), 0.995 (OFD), 0.875 (Atrial Width), 0.975 (TCD), 0.918 (CMS), 0.891 (NFT), 0.886 (CC length), 0.821 (CC width), 0.849 (Vermis anteroposterior), 0.969 (Vermis superoinferior). When the reader panel used the semi-automated caliper placement by OHEA, the average (across all measurements) inter-rater variability in the measurements decreased (compared to manual caliper placement) by 77%, indicating an improved consistency among the reader panel. Further, as a result of semi-automated caliper placement, the average time for each measurement was reduced by 45%, and the number of keystrokes was reduced by 43%.
Conclusion
The precise caliper placement is crucial to assist novice users in the reliable assessment of fetal growth and neurodevelopment, essential for screening fetal central nervous system anomalies. By reducing the inter-rater variability through AI assistance, centers can benefit from improved consistency and standardization in clinical practice, resulting in higher quality care. The workflow benefits such as reduction in measurement times and keystrokes can potentially make prenatal ultrasound examinations faster, reduce operator fatigue, improve productivity, and reduce patient waiting times in high-volume centres.
Note: Origin Health Examination AssistantTM (OHEA) is now Origin Medical EXAM ASSISTANTTM (OMEA).

Purpose
Ventriculomegaly (dilated fetal cerebral ventricles) is a relatively common finding on prenatal ultrasound and can be considered a soft antenatal marker requiring a specialist referral for a detailed search of associated anomalies. We propose a deep learning system (DLS) for the automated quantification and screening of suspected ventriculomegaly to assist operators to provide timely referrals.
Methods
We obtained retrospective ultrasound (US) examinations of 298 mid-trimester pregnancies (normal [N], unilateral ventriculomegaly [VM]: 259/39) from 2 tertiary referral centers. On 514 2D US images deemed clinically appropriate by fetal medicine specialists (FMS), we trained (ground-truth: FMS caliper points) a DLS to automatically predict the caliper points for measuring the atrial width (AW) of the lateral ventricles. The predicted AW measurements were then classified into normal or suspected VM based on clinical guidelines (ISUOG). The suspected VM cases were further classified into prominent, mild, and severe categories. We assessed the DLS performance in the automated measurement (mean error [ME]) and screening (sensitivity [Sn], specificity [Sp], accuracy [Ac]; with 95% CI) by benchmarking against clinical gold-standard (FMS).
Results
On an independent test set of 226 images (186 cases), the MEs (in mm) in DLS AW measurements were 0.47+-0.56 (normal, 143 cases), 0.41+-0.37 (prominent, 18 cases), 0.71+-0.77 (mild, 20 cases), and 0.77+-0.97 (severe, 5 cases). Further, the normal and suspected VM cases were discriminated with a Sn, Sp, and Ac of 95.18% (92.82 - 97.53%), 95.74% (94.03 - 97.44%), and 95.53% (94.14 - 96.91%), respectively.
Conclusion
We successfully developed and validated a DLS for the automated quantification and screening of suspected VM cases. It’s clinical translation can help expecting mothers in low-resource and remote settings to receive timely referrals for detailed examination.
Limitations
Bilateral VM cases were excluded from the study. The study had a limited dataset size (only mid-trimester cases).
Ethics committee approval
This study received the IRB approval from both the tertiary centers, and data were anonymized (tenets of the Declaration of Helsinki).

.svg)